Xizhan Xu, Zezheng Gao, Fuquan Yang, Yingying Yang, Liang Chen, Lin Han, Na Zhao, Jiayue Xu, Xinmiao Wang, Yue Ma, Lian Shu, Xiaoxi Hu, Na Lyu, Yuanlong Pan, Baoli Zhu, Linhua Zhao, Xiaolin Tong*,Jun Wang*. Antidiabetic Effects of Gegen Qinlian Decoction via the Gut Microbiota are Attributable to Its Key Ingredient, Berberine.Genomics, Proteomics & Bioinformatics. In press
Wei Zheng, Yue Ma, Ai Zhao, Tingchao He, Na Lyu,Ziqi Pan, Geqi Mao, Yan Liu, Jing Li, Peiyu Wang,Jun Wang*, Baoli Zhu* & Yumei Zhang*. Compositional and functional differences in human gut microbiome with respect to equol production and its association with blood lipid level: a cross-sectional study.Gut Pathogens2019, 11:20 doi:10.1186/s13099-019-0297-6
Jun Wang#, Alexander Kurilshikov, Djawad Radjabzadeh, Williams Turpin, Kenneth Croitoru, Marc Jan Bonder, Matthew A. Jackson, Carolina Medina-Gomez, Fabian Frost, Georg Homuth, Malte Rühlemann, David Hughes, Han-na Kim, MiBioGen Consortium Initiative, Tim D. Spector, Jordana T. Bell, Claire J. Steves, Nicolas Timpson, Andre Franke, Cisca Wijmenga, Katie Meyer, Tim Kacprowski, Lude Franke, Andrew D. Paterson, Jeroen Raes, Robert Kraaij, Alexandra Zhernakova. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative.Microbiome, 2018 6:101
Jun Wang#, Liang Chen, Na Zhao, Xizhan Xu, Yakun Xu, Baoli Zhu. Of genes and microbes: solving the intricacies in host genomes.Protein Cell. 2018 9(5):446-461
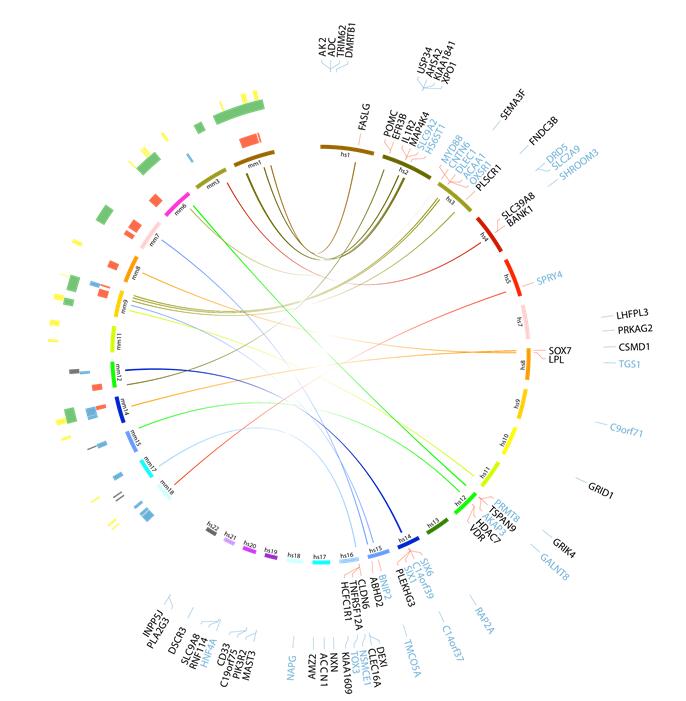
Jun Wang*, Louise B. Thingholm , Jurgita Skiecevi?ien?, *, Philipp Rausch, Martin Kummen, Johannes R. Hov, Frauke Degenhardt, Femke-Anouska Heinsen, Malte C. Rühlemann, Silke Szymczak, Kristian Holm, T?nu Esko, Jun Sun, Mihaela Pricop-Jeckstadt, Samer Al-Dury, Pavol Bohov, J?rn Bethune, Felix Sommer, David Ellinghaus, Rolf K. Berge, Matthias Hübenthal, Manja Koch, Karin Schwarz, Gerald Rimbach, Patricia Hübbe, Wei-Hung Pan, Raheleh Sheibani-Tezerji, Robert H?sler, Philipp Rosenstiel, Mauro D’Amato, Katja Cloppenborg-Schmidt, Sven Künzel, Matthias Laudes, Hanns-Ulrich Marschal, Wolfgang Lieb, Ute N?thlings, Tom H. Karlsen, John F. Baines, Andre Franke. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota.Nature Genetics, 2016 (48)1396–1406 (* contributed equally; Nature Genetics封面文章)
Gwen Falony*, Marie Joossens*, Sara Vieira-Silva*,Jun Wang*, Youssef Darzi, Karoline Faust, Alexander Kurilshikov, Marc Jan Bonder, Mireia Valles-Colomer, Doris Vandeputte , Raul Y Tito, Samuel Chaffron, Leen Rymenans, Chlo? Verspecht, Lise De Sutter, Gipsi Lima-Mendez, Kevin D'hoe, Karl Jonckheere, Daniel Homola, Roberto Garcia, Ettje F. Tigchelaar, Linda Eeckhaudt, Jingyuan Fu, Liesbet Henckaerts, Alexandra Zhernakova, Cisca Wijnenga, Jeroen Raes. Population-level analysis of gut microbiome variation.Science, 2016, 352(6285):560-565 (* contributed equally; Science封面文章)
Alexander R. Moschen*, Romana R. Gerner*,Jun Wang*, Victoria Klepsch, Timon E. Adolph, Simon J. Reider, Hubert Hackl, Alexandra Pfister, Johannes Schilling, Patrizia L. Moser, Sarah L. Kempster, Alexander Swidsinski, Dorothea Orth?H?ller, Günter Weiss, John F. Baines, Arthur Kaser, Herbert Tilg. Lipocalin 2 protects from colonic inflammation and tumorigenesis through its microbiota modulating properties.Cell Host Microbes, 2016, 19(4):455–469 (* contributed equally)
Jun Wang, Shirin Kalyan, Natalie Steck, Leslie M. Turner, Bettina Harr, Sven Künzel, Marie Vallier, Robert H?sler, Andre Franke, Hans-Heinrich Oberg, Saleh M. Ibrahim, Guntram A. Grassl, Dieter Kabelitz, John F. Baines. Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome.Nature Communications2015, 6:6440.
Jun Wang, Miriam Linnenbrink, Sven Künzel, Ricardo Fernandes, Marie-Josée Nadeau, Philip Rosenstiel, John F Baines. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice.Proceedings of the National Academy of Sciences2014, 11(26):E2703-E2710.
Miriam Linnenbrink*,Jun Wang*, Emilie A Hardouin, Sven Künzel, Dirk Metzler, John F Baines: The role of biogeography in shaping diversity of the intestinal microbiota in house mice.Molecular Ecology2013, 22(7):1904-1916.