2. Cui YL, Chen YC, Liu XY, Dong SJ, Tian YE, Qiao YX, Mitra R, Han J, Li CL, Han X, Liu WD, Chen Q, Wei WQ, Wang X, Du, Tang SY, Xiang H, Liu HY, Liang Y, Houk KN,Wu B*. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy.ACS Catal.2021, 11(3): 1340–1350. doi: 10.1021/acscatal.0c05126.(封面文章)
3. Sun JY,CuiLY,Wu B*. GRAPE, a greedy accumulated strategy for computational protein engineering.Meth. Enzymol.2021,648:207-230. doi: 10.1016/bs.mie.2020.12.026.
4. Zhu T, Li RF, Sun JY, Cui YL,Wu B*. Characterization and efficient production of a thermostable, halostable and organic solvent-stable cellulase from an oil reservoir.Int. J. Biol. Macromol.2020, 159: 622-629. doi: 10.1016/j.ijbiomac.2020.05.021.
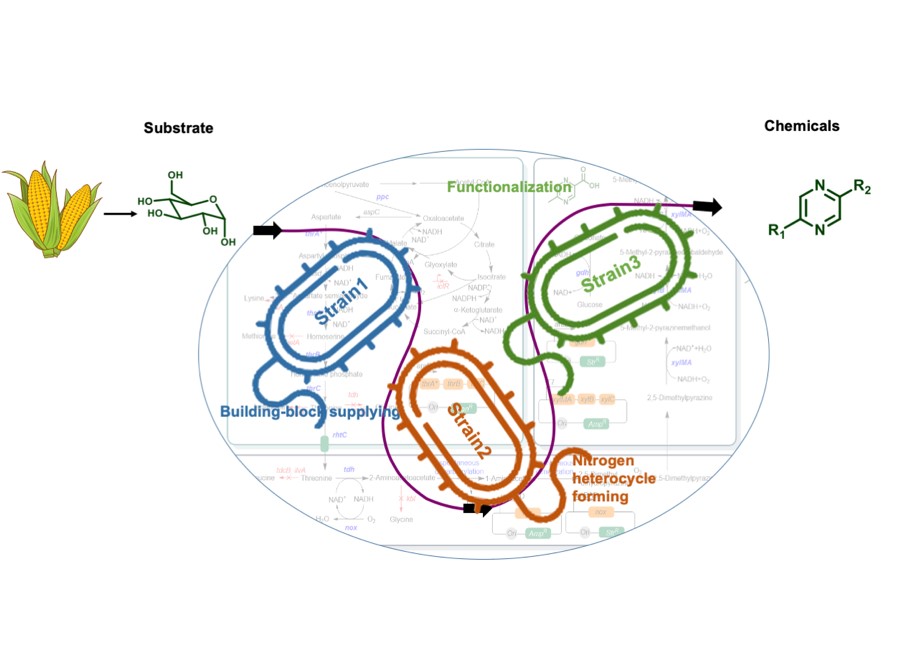
5. Feng J, Li RF,Zhang SS, Bu YF, Chen YC, Cui YL,Lin BX, Chen YH, Tao Y,Wu B*. Bioretrosynthesis of Functionalized N-Heterocycles from Glucose via One-Pot Tandem Collaborations of Designed Microbes.Adv.Sci.2020,7, 2001188.doi: 10.1002/advs.202001188.
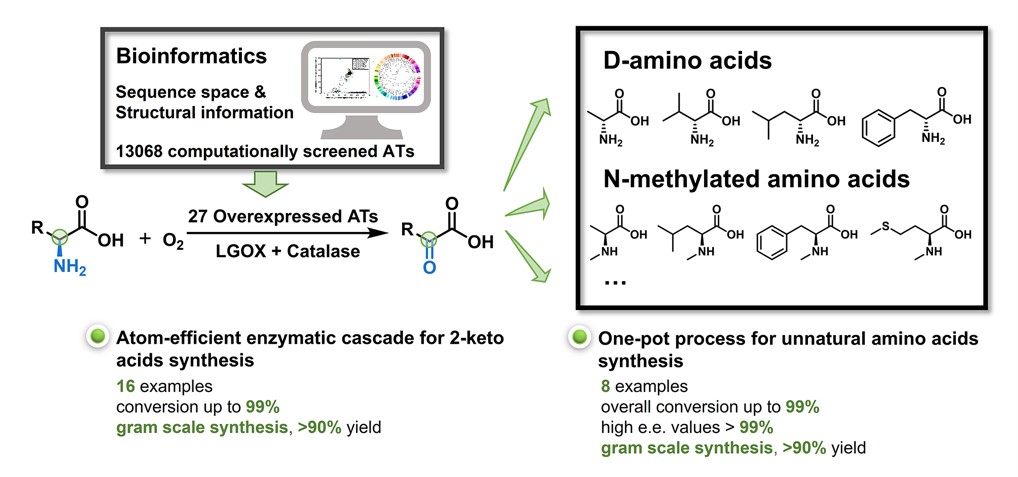
6. Li T,Cui XX,Cui YL, Sun JY, Chen YC,Zhu T,Li CJ, Li RF,Wu B*. Exploration oftransaminasediversity for theoxidativeconversion ofnaturalaminoacids into 2-ketoacids andhigh-valuechemicals.ACS.Catal.2020,10(14), 7950–7957.doi: 10.1021/acscatal.0c01895.
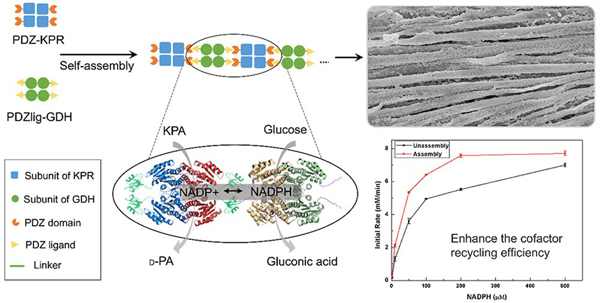
7. Li T, Li RF, Zhu T, Cui XX, Li CJ, Cui YL,Wu B*. Improving the system performance of the asymmetric biosynthesis of D-pantoic acid by using artificially self-assembled enzymes inEscherichia coli.ACS.Biomater. Sci. Eng.2020, 6(1) , 219–224. doi: 10.1021/acsbiomaterials.9b01754.
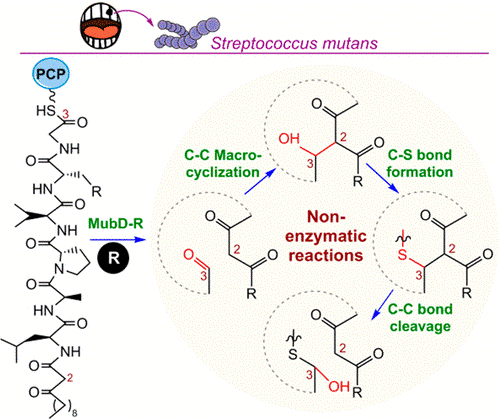
8. Wang M, Xie ZJ, Tang SB, Chang EL, Tang Y, Guo ZG, Cui YL,Wu B, Ye T, Chen YH. Reductase of mutanobactin synthetase triggers sequential C–C macrocyclization, C–S bond formation, and C–C bond cleavage.Org. Lett.2020, 22(3) , 960–964. doi: 10.1021/acs.orglett.9b04501.
9. Mu QX, Cui YL, Tian YE, Hu MR, Tao Y,Wu B*. Thermostability improvement of the glucose oxidase fromAspergillus nigerfor efficient gluconic acid production via computational design.Int. J. Biol. Macromol.2019, 136: 1060-1068. doi: 10.1016/j.ijbiomac.2019.06.094.
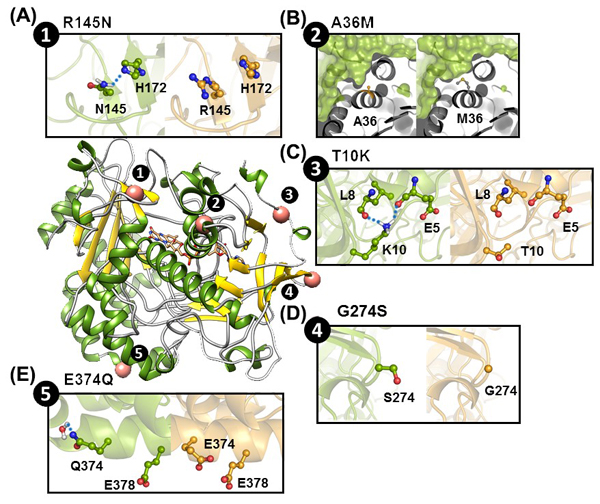
10. Zheng XX, Cui YL, Li T, Li RF, Guo L, Li DF,Wu B*. Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase fromPseudomonassp. for efficient biosynthesis of chiral amino acid.Appl. Microbiol. Biotech.2019, 103(19): 8051-8062. doi: 10.1007/s00253-019-10105-9.
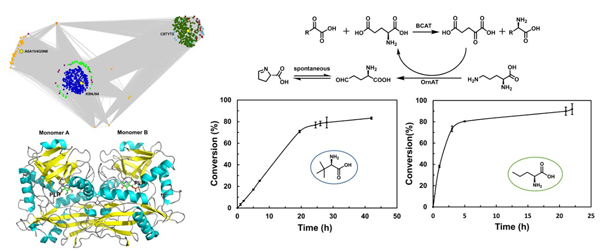
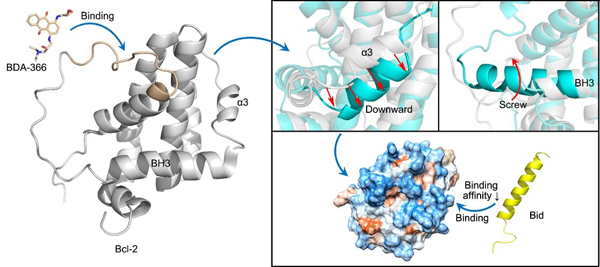
11. Li T, Cui YL,Wu B*. Molecular dynamics investigations of structural and functional changes in Bcl-2 induced by the novel antagonist BDA-366.J. Biomol. Struct. & Dyn.2019, 37(10): 2527-2537. doi:10.1080/07391102.2018.1491424.
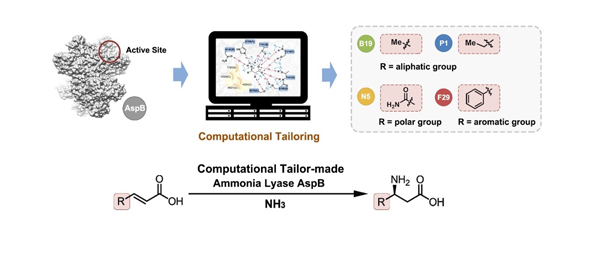
12. Li RF, Wijma HJ, Song L, Cui YL, Otzen M, Tian YE, Du JW, Li T, Niu DD, Chen YC, Feng J, Han J, Chen H, Tao Y, Janssen DB*,Wu B*. Computational redesign of enzymes for regio- and enantioselective hydroamination.Nat. Chem. Biol.2018, 14(7): 664-670. doi: 10.1038/s41589-018-0053-0.
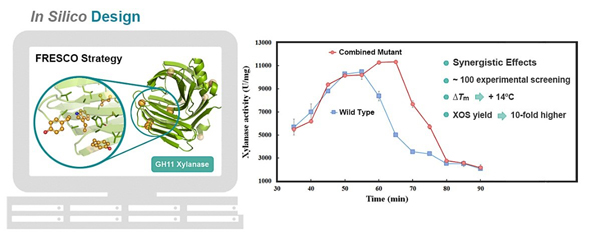
13. Bu YF, Cui YL, Peng Y, Hu MR, Tian Y’E, Tao Y,Wu B*. Engineering improved thermostability of the GH11 xylanase fromNeocallimastix patriciarumvia computational library design.Appl. Microbiol. Biotech.2018, 102(8): 3675-3685. doi: 10.1007/s00253-018-8872-1.
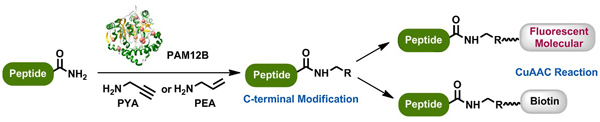
14. Zhu T, Song L, Li RF,Wu B*. Enzymatic clickable functionalization of peptides via computationally engineered peptide amidase.Chin. Chem. Lett.2018, 29(7): 1116-1118. doi: 10.1016/j.cclet.2018.03.033.
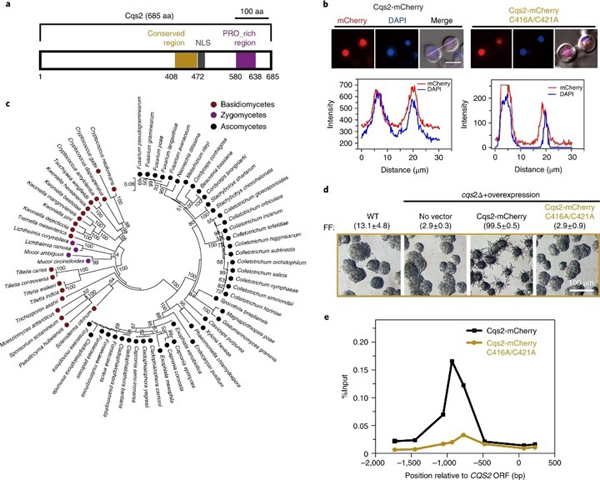
15. Tian XY, He GJ, Hu PJ, Chen L, Tao CY, Cui YL, Shen L, Ke WX, Xu HJ, Zhao YB, Xu QJ, Bai FY,Wu B,Yang E, Lin XR, Wang LQ*.Cryptococcus neoformanssexual reproduction is controlled by a quorum sensing peptide.Nat. Microbiol.2018, 3(6): 698-707. doi: 10.1038/s41564-018-0160-4.
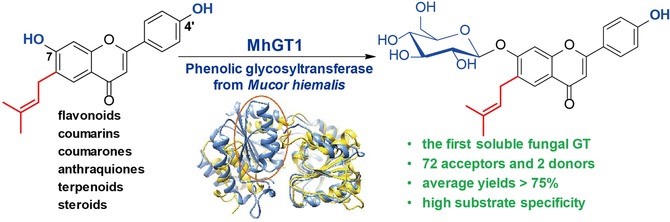
16. Feng J, Zhang P, Cui YL, Li K, Qiao X, Zhang YT, Li SM, Cox R.J,Wu B, Ye M*, Yin WB*. Regio-and stereospecificO-glycosylation of phenolic compounds catalyzed by a fungal glycosyltransferase fromMucor hiemalis.Adv. Synth. Catal.2017, 359(6): 995-1006. doi: 10.1002/adsc.201601317.
17. Wu B*, Wijma HJ, Song L, Rozeboom HJ, Poloni C, Tian YE, Arif MI, Nuijens T, Quaedflieg PJ, Szymanski W, Feringa BL, Janssen DB*. Versatile peptide C-terminal functionalization via a computationally engineered peptide amidase. ACS.Catal.2016, 6(8): 5405-5414. doi: 10.1021/acscatal.6b01062.
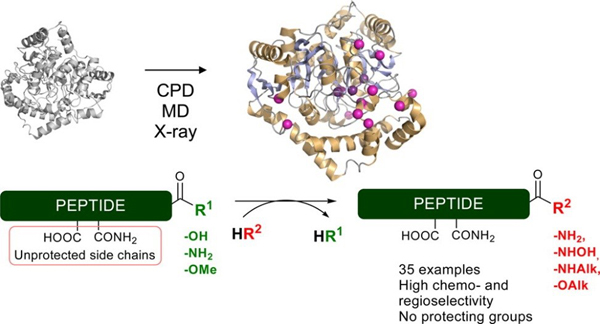
18. Toplak A, Nuijens T, Quaedflieg PJ, Wu B*, Janssen DB*. Peptiligase, an enzyme for efficient chemoenzymatic peptide synthesis and cyclization in water.Adv. Synth. Catal.2016, 358(13): 2140-2147. doi: 10.1002/adsc.201600017.
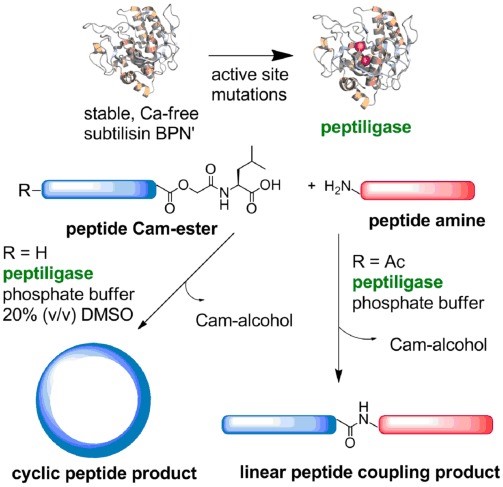
19. Nuijens T , Toplak A , Quaedflieg PJLM , Drenth J ,Wu B*,Janssen DB*. Engineering a diverse ligase toolbox for peptide segment condensation.Adv. Synth. Catal.2016, 358(24): 4041-4048. doi: 10.1002/adsc.201600774.